Sex chromosome aneuploidies, conditions where individuals have extra or missing X or Y chromosomes, are among the most frequent chromosomal abnormalities detected in prenatal and postnatal settings. From Turner syndrome (45,X) to Klinefelter syndrome (47,XXY), accurately and quickly identifying these aneuploidies is vital for genetic counseling and informed medical decision-making.
In this article, we walk through how QF-PCR (Quantitative Fluorescent PCR), using the Devyser Complete v2 kit. But before we dive into the specific aneuploidies, let’s first understand the tools we’re using.
What is QF-PCR?
QF-PCR is a technique that amplifies specific regions of DNA, mostly short tandem repeats (STRs), and quantifies them using fluorescent signals. This allows us to estimate how many copies of a chromosome are present in the sample.
QF-PCR is especially suited for detecting trisomies (three copies of a chromosome) or monosomies (only one copy). In the case of sex chromosomes, it can distinguish between normal karyotypes and abnormalities like Turner (45,X), Klinefelter (47,XXY), XYY, or Triple X (47,XXX).
A Brief Overview of the Protocol (Devyser Complete v2)
- DNA Extraction: DNA is extracted from blood, amniotic fluid, or chorionic villus sample.
- PCR Amplification: The Devyser kit includes two multiplex mixes (Mix 1 and Mix 2), each containing primers targeting STRs and key non-polymorphic markers.
- Capillary Electrophoresis: Amplified DNA is run on an instrument like the ABI 3500. Fluorescent peaks are recorded.
- Data Analysis: The pattern and peak ratios (e.g. 1:1, 2:1, 3:1) are analyzed using software (e.g., GeneMapper). These patterns help determine if there’s a normal or abnormal number of chromosomes.
Key Markers Used in Sex Chromosome Analysis
Let’s break down the core markers that help identify sex chromosome composition:
1. AMELXY (Amelogenin gene)
- Found on both X and Y chromosomes, but the PCR product sizes are different.
- X = 104 bp, Y = 110 bp
- If both peaks are present → male
- If only the X peak is present → female
Purpose: Confirms the presence or absence of the Y chromosome and supports sex determination.
2. SRY (Sex-determining Region Y)
- A non-polymorphic Y-specific marker
- Present only in males (on the Y chromosome)
- Strong confirmatory evidence for the presence of a Y chromosome
Purpose: Crucial in detecting Y material, especially in Turner variants or disorders of sex development (DSDs).
3. T1 and T3: X-Counting Markers
- These markers compare a non-variable (non-polymorphic) sequence on the X chromosome with a similar sequence on an autosome (Chromosome 7 for T1, Chromosome 3 for T3).
- Both regions are amplified with the same primers, but result in different-sized PCR products (X vs autosome).
- The peak area ratio (X:autosome) indicates the number of X chromosomes:
| Ratio | Interpretation |
|---|---|
| 1:1 | Two Xs (normal female) |
| 0.5:1 | One X (Turner or normal male) |
| 1.5:1 | Three Xs (Triple X or XXY) |
Purpose: Provides a consistent internal control for X chromosome dosage estimation.
4. X-STR Markers (e.g., DXS1187, DXS981)
- STRs (short tandem repeats) located on the X chromosome.
- These are polymorphic — their length varies between individuals.
- Can show one peak (in males or Turner) or two peaks (in females), depending on zygosity.
- In cases of extra X chromosomes, may show three peaks or peak ratios like 2:1.
Purpose: Reveals both X chromosome copy number and heterozygosity.
Now that we understand the tools, let’s walk through how each sex chromosome aneuploidy appears on QF-PCR.
Commonly Encountered QF-PCR Patterns
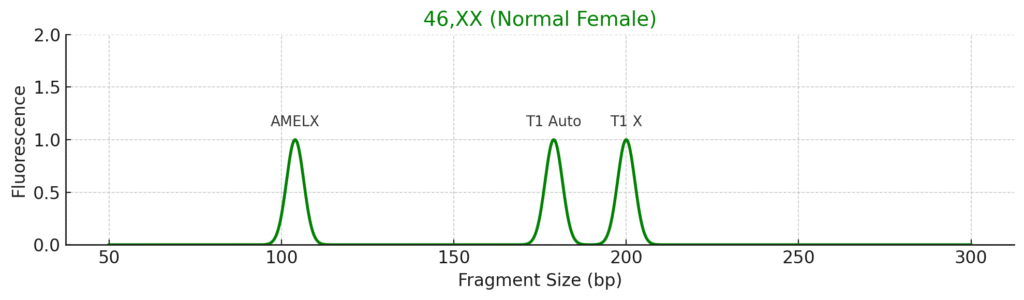
1. Normal Female (46,XX)
- AMELXY: Only AMELX (104 bp)
- SRY: Absent
- T1/T3: ~1:1 ratio → two X chromosomes
- X-STRs: Two peaks (if heterozygous), 1:1 ratio
- XY-STRs: Absent or shows X alleles only
Conclusion: Normal female karyotype
2. Normal Male (46,XY)
- AMELXY: AMELX + AMELY
- SRY: Present
- T1/T3: ~0.5 ratio → one X chromosome
- X-STRs: One peak (hemizygous)
- XY-STRs: Two peaks (X and Y alleles)
Conclusion: Normal male karyotype
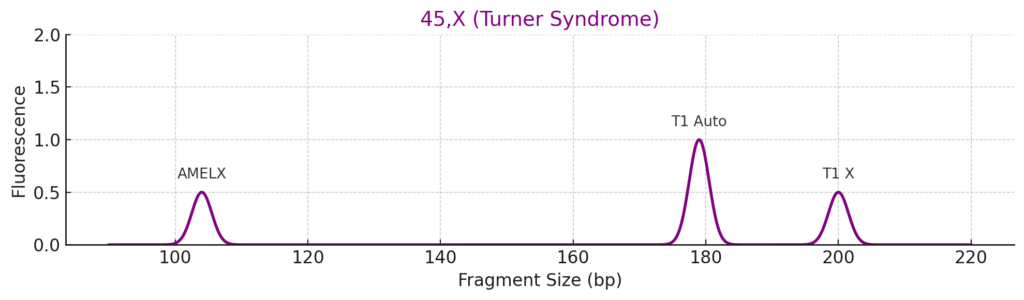
3. Turner Syndrome (45,X)
- AMELXY: Only AMELX
- SRY: Absent
- T1/T3: ~0.5 → one X chromosome
- X-STRs: Single peak for each marker (homozygous)
- XY-STRs: Absent or only X
Conclusion: Monosomy X — Turner syndrome
4. Klinefelter Syndrome (47,XXY)
- AMELXY: AMELX + AMELY
- SRY: Present
- T1/T3: 1.0 → two X chromosomes
- X-STRs: Either 3 peaks or 2 peaks with skewed ratio (2:1)
- XY-STRs: Show X and Y alleles
Conclusion: Two Xs and one Y — Klinefelter syndrome
5. XYY Syndrome (47,XYY)
- AMELXY: X + stronger Y peak (AMELY)
- SRY: Present (often stronger)
- T1/T3: ~0.5 (one X chromosome)
- X-STRs: One peak (hemizygous)
- XY-STRs: Skewed patterns due to two Ys
Conclusion: One X and two Y chromosomes — XYY syndrome
Summary Table
| Condition | AMELXY | SRY | T1/T3 Ratio | X-STRs | Interpretation |
|---|---|---|---|---|---|
| 46,XX (Female) | X only | Absent | ~1:1 | 2 peaks (1:1) or 1 peak (homozygous) | Normal female |
| 46,XY (Male) | X + Y | Present | ~0.5:1 | 1 peak (hemizygous) | Normal male |
| 45,X (Turner) | X only | Absent | ~0.5:1 | 1 peak | Turner syndrome |
| 47,XXY | X + Y | Present | ~1:1 | 2 peaks (1:1) or 1 peak (homozygous) | Klinefelter syndrome |
| 47,XYY | X + strong Y | Present | ~0.5:1 | 1 peak | XYY syndrome |
| 47,XXX | X only | Absent | ~1.5:1 | 3 peaks or 2:1 | Triple X syndrome |
Conclusion
QF-PCR offers a quick, accurate, and cost-effective way to diagnose sex chromosome aneuploidies. By understanding the roles of polymorphic STRs and non-polymorphic markers like AMELXY, SRY, T1, and T3, clinicians and geneticists can confidently interpret complex results with high resolution with quick turnaround time.
Join Our Google Group
Join our google group and never miss an update from Gene Commons.